Degnan lab > Crown of thorns starfish
Crown-of-thorns starfish (COTS) genome
Acanthaster planci
- Animal communication in the ocean
- Genome-guided biocontrol
Acanthaster planci is a highly fecund predator of reef-building corals throughout the Indo-Pacific region. Population outbreaks of this starfish cause substantial loss of coral cover, diminishing the integrity and resilience of reef ecosystems. The genome from a Great Barrier Reef COTS provides insights into conspecific communication that may guide the generation of biocontrol mimetics for use on reefs with COTS outbreaks.
More than 150 transcriptomes from Great Barrier Reef COTS that allow for the analysis of gene activity between sexes of wild COTS across breeding and non-breeding seasons, between multiple discrete tissues and organs, and when starfish are translocated from the wild into captivity.
COTS Browser guide
The COTS Apollo browser allows visualisation of gene models and of mapped RNA-Seq and CEL-Seq2 reads. For general navigation of the Apollo browser, we refer you to the Apollo user guide.
The v1.1 PASA gene models are shown by default on the COTS browser. Below is an example view of the browser with two mapped transcriptomes.
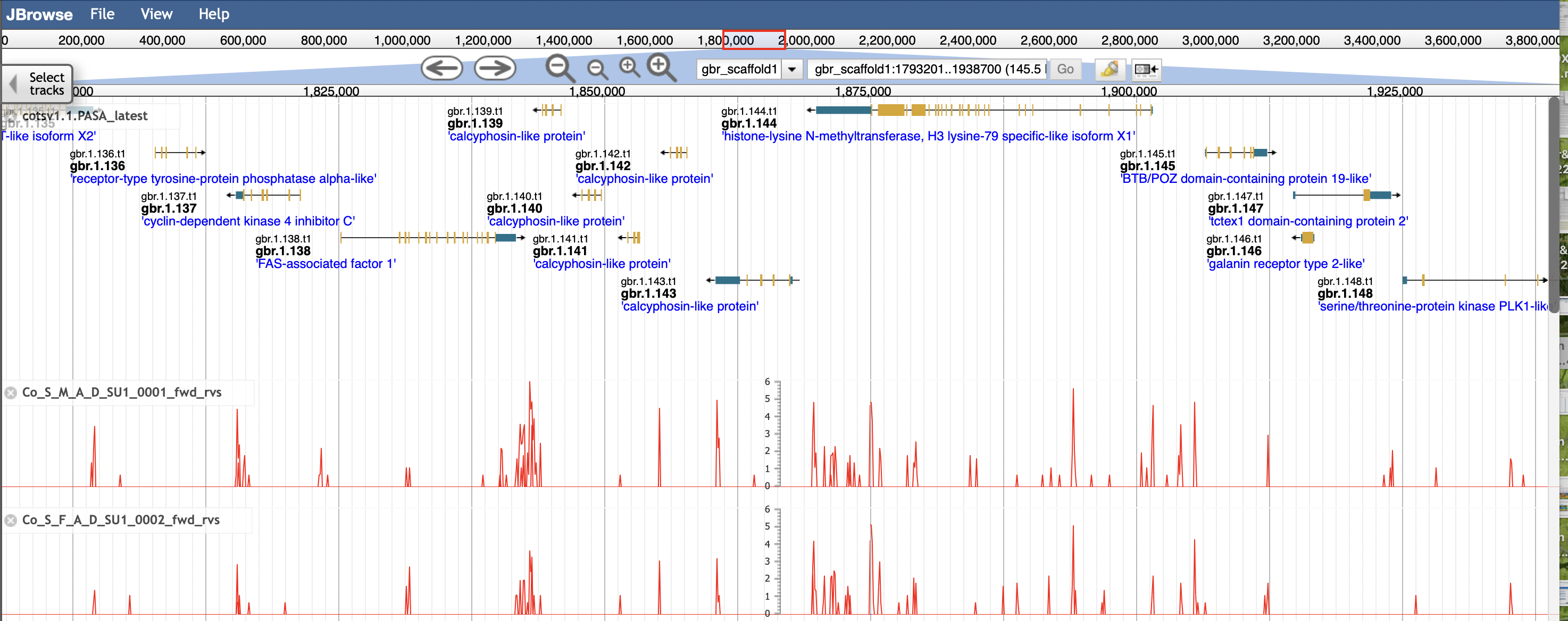
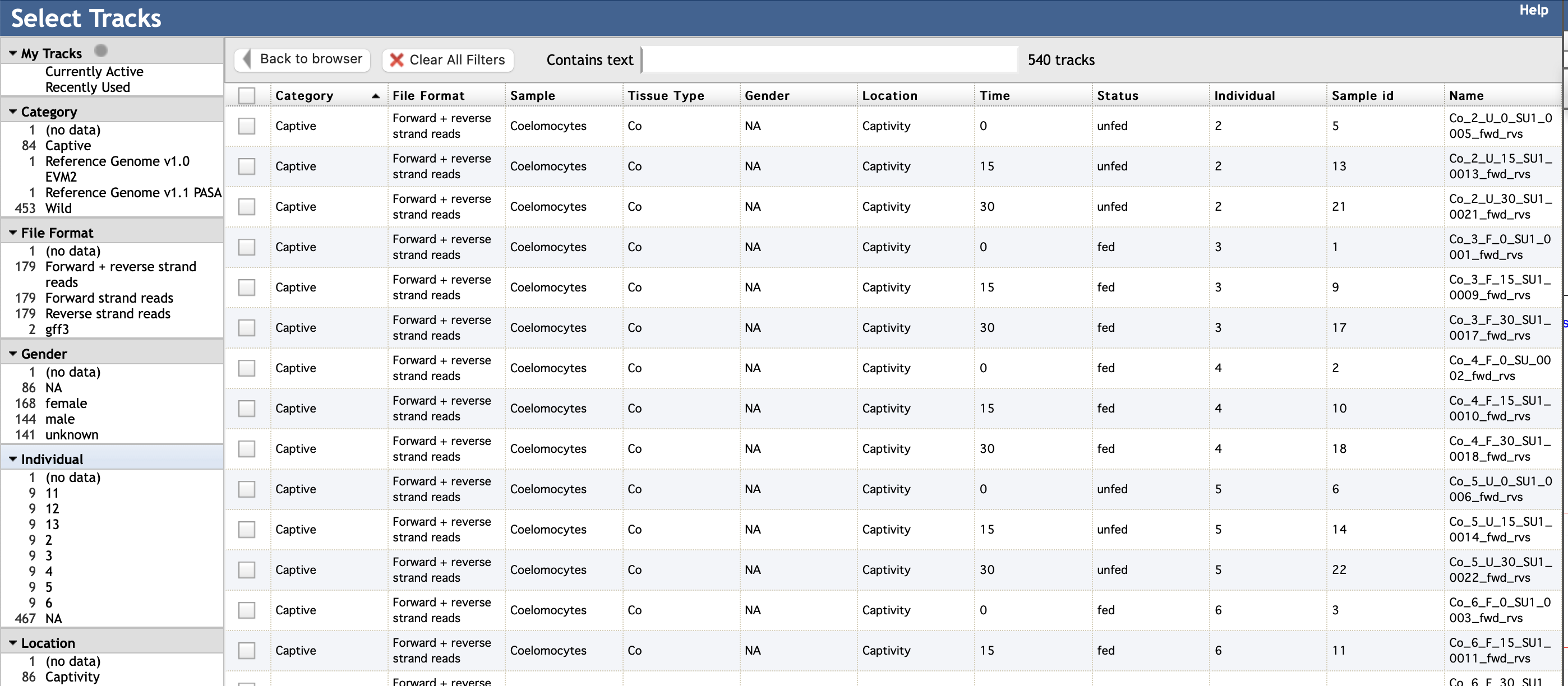
You can select to view older gene models and specific transcriptomes by clicking on the
Select tracks tab in the top left corner. Here you can select specific tracks
(transcriptomes and gene model versions) and how you would like to view them. It looks
like this:
You can select specific elements in the left column according to your interest. For instance, to view CEL-Seq2 reads mapping to forward and reverse strands of the genome from transcriptomes made from gonads of wild female COTS sampled from Davies Reef (on the Great Barrier Reef), you would select the following elements (in blue):
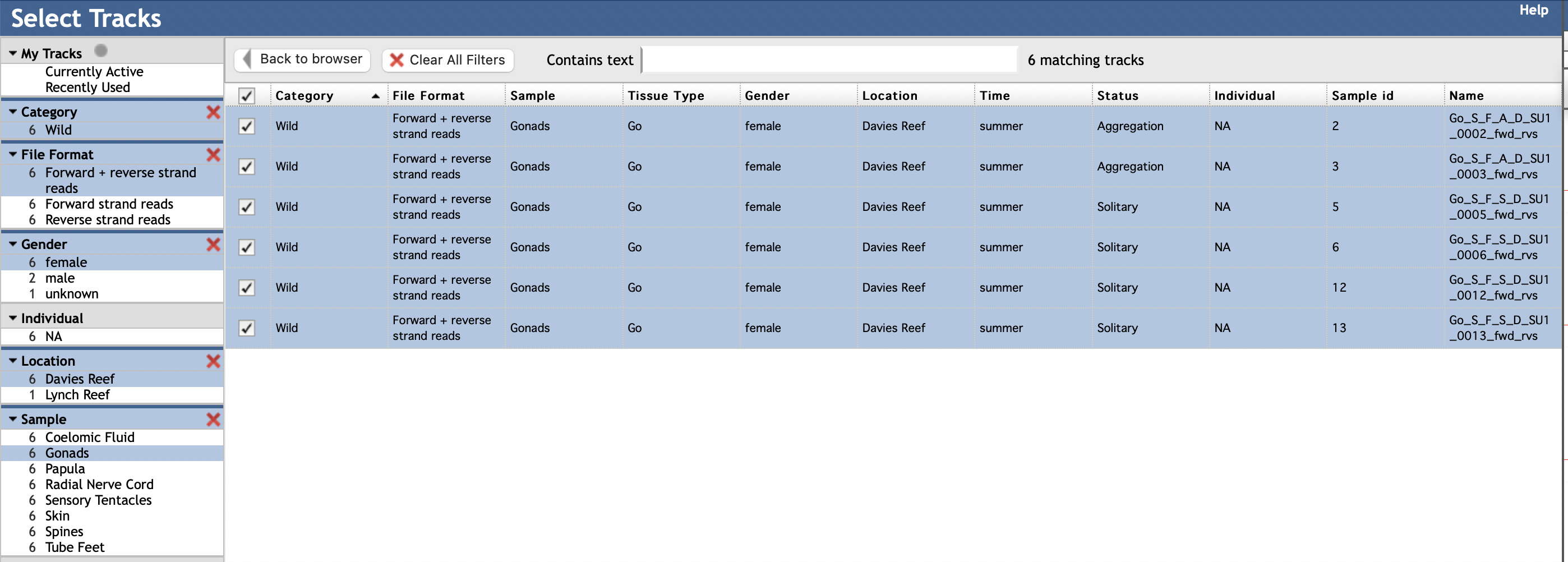
You can add or subtract from this list by selecting and deselecting individual elements or groups of elements. If you wish to create a new set of tracks, it is best to refresh the browser to clear your previous views.
Transcriptome track fields
Transcriptomes are grouped and identified by the following fields:
Category
- Captive - COTS maintained in captivity
- Wild - COTS sampled in the field
File format - BAM files to show RNA-Seq reads mapped to
- Both forward and reverse strands of the genome
- Forward strand only
- Reverse strand only
Sample and tissue/organ type
- Coelomocytes (Co)
- Gonads (Go)
- Papulae (Pa)
- Radial nerves (Rn)
- Sensory tentacles (St)
- Skin (Sk)
- Spines (Sp)
- Tube feet (Tf)
Sex
- Male
- Female
- Unknown - animals whose sex could not be determined because of season or age (i.e. non-reproductive)
Location: either captivity or the specific reef of capture.
Time: either specific season of collection, or experimental time point
Status: related to observation at time of field collection relative to other COTS, or type of experimental treatment. See relevant literature for details.
Individual & Sample ID: corresponds to individual COTS and tissues sampled, and matches transcriptome identifiers in NCBI BioProject PRJNA821257.
Name: combined name of specific transcriptome. This is listed to the left of the browser and corresponds to the NCBI BioProject identifier.
Funding
We would like to acknowledge the following funding bodies for their support of this project


